Metagenomics Services
Frontage offers advanced sequencing technologies that enable profiling of the microbiome DNA composition (metagenomics) and the microbiome RNA composition (metatranscriptomics) at unprecedented scales allowing for the discovery of invaluable taxonomic and functional profiles of the microbial community.
Metagenomics
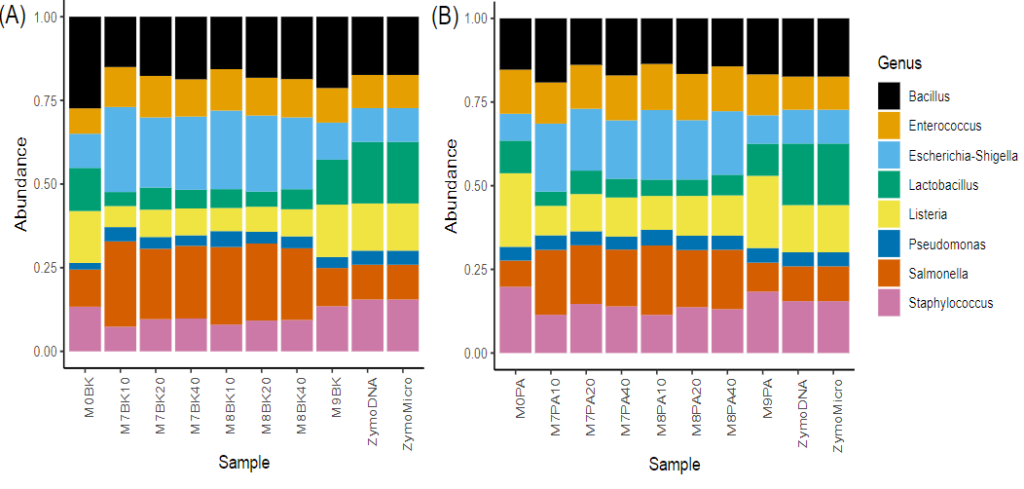
Metagenomics is the study of DNA sequences comprising a population of organisms making up specific environmental niches including specific soil types, water samples, or human skin or gut microflora. Frontage offers sequencing with primer sets that span both V3 and V4 HVRs which have been recommended by Illumina (BK primer set), or an alternative set that spans V4 only and which has been utilized by the Earth Microbiome Project (PA primer set), the latter having been optimized for detection of Archaea. Both primers sets have been validated in Frontage’s laboratory and typical results are shown in Figure 1.
Frontage can also implement custom primer sets for clients with unique experimental goals.
Frontage’s metagenomic analysis service starts with DNA isolation using the ZymoBIOMICS DNA/RNA Miniprep kit which is designed for uniform and efficient lysis of many different organisms (e.g. Gram-negative/positive bacteria, fungus, protozoans, and algae) from a variety of sample inputs. Next the V3-V4 or V4 regions of the 16S rDNA are amplified with either the Bakt or Parada primer sets. The 16S rDNA amplicons are then adapted with Illumina-instrument compatible PCR priming sites via an additional 8 cycles of PCR. Amplicons are sequenced on the MiSeq or HiSeq 2500 sequencing instruments from Illumina. For more information on this, read our white paper: 16S rDNA Amplicon Sequencing of Mock Microbial Community Standards.
We perform comprehensive metagenomic analyses for elucidating taxonomic diversity and phylogenetic relationships within various sample types.
Metatranscriptomics
Frontage’s metatranscriptomics services facilitate gene expression studies on a global scale, revealing the transcriptionally active portion of the microbiome, i.e., the portion that contributes to the functional capacity of the microbiome. Our metatranscriptome services are offered for all stages of your microbiomics project. We perform RNA extraction, library preparation, sequencing, and analysis of your complex metatranscriptomic data including interactive taxonomic classification reports, as well as gene-level expression analysis and visualization
Metatranscriptomics can be used in multiple areas of research:
- Determine drug treatment responses with measurable changes in microflora composition and gene expression
- Microflora RNA signatures can be associated with specific-disease subtypes and enable patient stratification, including identification of non-responders.
- Study induction or repression of specific metabolic pathways in response to changes in diet or during disease development.
- General mechanism of action studies.
- Improve diagnostics for infectious disease status and susceptibility.
- Study interactions between symbiotic bacteria and host.
Sample Processing for Metatranscriptomic Analysis
Frontage has developed well-validated protocols for the extraction of RNA from common and unique biospecimens. Examples of accepted sample types are given below, but Frontage has the capabilities to work with many unique sample types.
- Stool / Feces
- Tissue biopsies
- Cells
- Saliva
- Bronchial lavage
- Sputum
- Buccal swabs
- Oral biofilms
- Soil
- Plants
- Food items
Frontage has the latest technology from Illumina to perform high coverage sequencing (NextSeq 550Dx, NextSeq 2000).
Microbiome Services
Our scientific team has over a decade of experience in helping sponsors solve the development problems of therapeutics, probiotics and prebiotics.
Learn MoreRNA Sequencing Services
Our RNA sequencing services use a combination of state-of-the-art equipment that provides quick and accurate results for your RNA profiling projects.
Learn More